Mapping membrane activity using machine learning
PNAS 2016
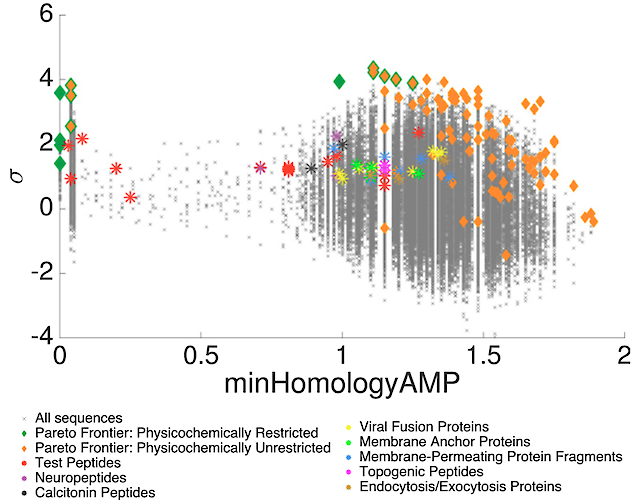
We trained a machine-learning classifier to identify membrane active alpha-helical peptides capable of remodeling and penetrating cell membranes. Classifier predictions were validated against calibrating small angle X-ray scattering measurements and deployed to perform high-throughput virtual screening to prospectively discover novel membrane-active sequences. We discover a diverse taxonomy of membrane-active peptides within differentiated protein families, including neuropeptides, viral fusion proteins, and amyloids, and design novel membrane-active peptides that are mutationally distant from any natural sequences. The classifier was intentionally designed to be human interpretable to reveal a comprehensible physicochemical basis for the topological basis for membrane activity and furnish actionable design rules for de novo membrane active peptide design. This work provides the foundations for the interpretation and extraction of design rules for protein function from learned latent space representations.